LiveDocs of Granule Cells Analysis.
Click here to open interactive Notebook (GWDG hosted):
Click here to open interactive Notebook (Binder hosted):
DESCRIPTION
Notebooks
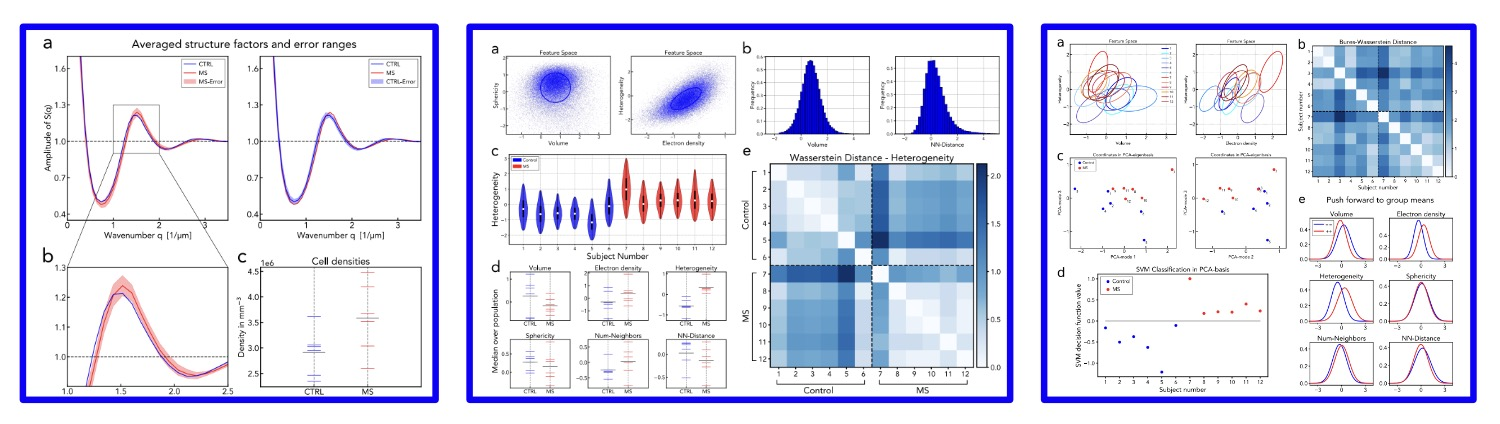
The notebook nb1_structure_factor.ipynb contains the calculation of the structure factors and the cell density of all patients out of the positions of the nuclei.
The notebook nb2a_prepare-data-and-1d-analysis.ipynb contains the creation of the feature space and the 1d histogram analysis (t-test, median values and 1dOT).
Furthermore, the Gaussian approximations and the linear embedding are calculated for the multidimensional analysis with OT (see nb2b_OT_analysis.ipynb).
The notebook nb2b_OT_analysis.ipynb builds on the previous nb2a_prepare-data-and-1d-analysis.ipynb notebook and contains the multidimensional analysis with Optimal Transport
with all results up to the final push forwards.
Folders
The folder Data/txt-data contains txt-tables with the positions and properties of the nuclei obtained from the segmentation. Each file corresponds to one patient where the file-numbers indicate the patient numbers. Note this data is also provided in the excel-files.
The folder Data/Data-for-Transport-Analysis contains all the data created/intermediately stored in the script Prepare-data, and then used in the script Transport-Analysis (Gaussian approximations, linear embedding, reference sample )
The folder lib contains a collection of Optimal Transport Algorithms (Barycenter computation, OT Solver) created by Bernhard Schmitzer and the
Optimal Transport Group in Göttingen (unpublished). The documented library can be found also on Github: https://github.com/bernhard-schmitzer/UnbalancedLOT
The file Data/params.txt contains chosen parameters for the entropic regularized solver of the transport problem.
Credits / Citation
DOI: doi.org/10.1016/j.neuroscience.2023.04.002
Cite the manuscript below, when using these scripts or data.
3d virtual histology reveals pathological alterations of cerebellar granule cells in Multiple Sclerosis. Jakob Frost, Bernhard Schmitzer, Mareike Töpperwien, Marina Eckermann, Jonas Franz, Christine Stadelmann, Tim Salditt NeuroScience (2023).
In case of questions, please address authors of the corresponding manuscript.
